At Genesis and Genetics, we have examined Y-chromosome genomes searching for Noah and his three sons, Ham, Japheth, and Shem. They were easy to find. According to our analysis, if you have the rs17306671 Y-chromosome mutation nucleotide A you are from Shem. If you have the rs9786139 Y-chromosome mutation nucleotide A you are from Ham. If you have the rs3900 Y-chromosome mutation nucleotide G you are from Japheth. If you are a woman, you will have to check your father’s y-chromosome since women don’t have the y-chromosome.
The following presents the easy-to-follow logic and analysis which anyone can repeat. Our findings are consistent with the Bible and modern science raw data.
Here is a bit of background: Y-chromosome DNA is exclusively found in males; it is inherited from one’s father. The Y-chromosome has approximately 60 million base pairs, each of which is subject to mutations. Mutations develop in the Y-chromosome, typically at the rate of 2 mutations per generation. This is based on a mutation rate of 1.0*10^-9 mutations /nucleotide/year (Reference 1) and 30 years per generation. These mutations allow us to track ancestry. If one man populated all the earth, all males would have his Y-chromosome, and if this man had two sons, one would expect that roughly half of the world would have one son’s mutations and the other half would have the other son’s mutations. Furthermore, if our original ancestor had 10 grandsons, one would expect that each grandson would have mutations that would each exist in approximately 10 percent of the male population. Successive generations would continue to generate new mutations that would form a human family tree, known as a phylogenetic tree to geneticists.
In our research, we used the above principles in tracking our most ancient ancestors. We made no assumptions and shelved our preconceived ideas. We wanted to see where the data led us. Appendix A shows the precise method we used to extract the data. Our strategy was simple; we gathered the Y-chromosome mutations, which are seen in more than 5 percent of human males, and entered them into a spreadsheet. We looked for patterns and color-coded the resulting groups. Please consider doing one Y-chromosome example yourself using Appendix A as a guide, or if you have your Y-chromosome data, you can trace it back to your most ancient ancestor/ancestors. You will see the ancient ancestor to whom you are related.
The data used in this research came from the 1000 genomes project (Reference 2) and retrieved using the Ensembl browser (Reference 3). The data appeared to be correct with no errors. No data was eliminated due to suspect errors, and no data was “cherry-picked” to suit any preconceived ideas. Our thanks to the great effort of those who did the sequencing and publishing of the raw data. We also greatly appreciate that it was made available to the public.
We started by taking 57 Y-genomes of diverse people (Americas, East Asia, Europe, South Asia, and Africa). Next, we gathered mutations that were in at least 5 percent of the world’s male population. The technical term for this is those with a Mean Allele Frequency (MAF) greater than or equal to 0.05 (5 percent). These mutations are the most interesting; any smaller MAF mutation is a subset of those greater than 5%.
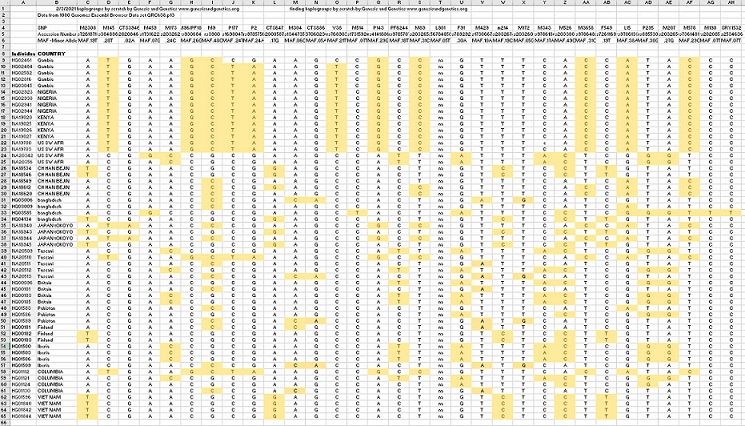
The spreadsheet below provides 57 rows (individual male humans) and 30 columns (mutations over 5 percent of the population). Each column element of the matrix was color-coded to show us whether the individual had the mutation or not. We noticed patterns beginning to form (you can CLICK HERE to see the EXCEL copy.)
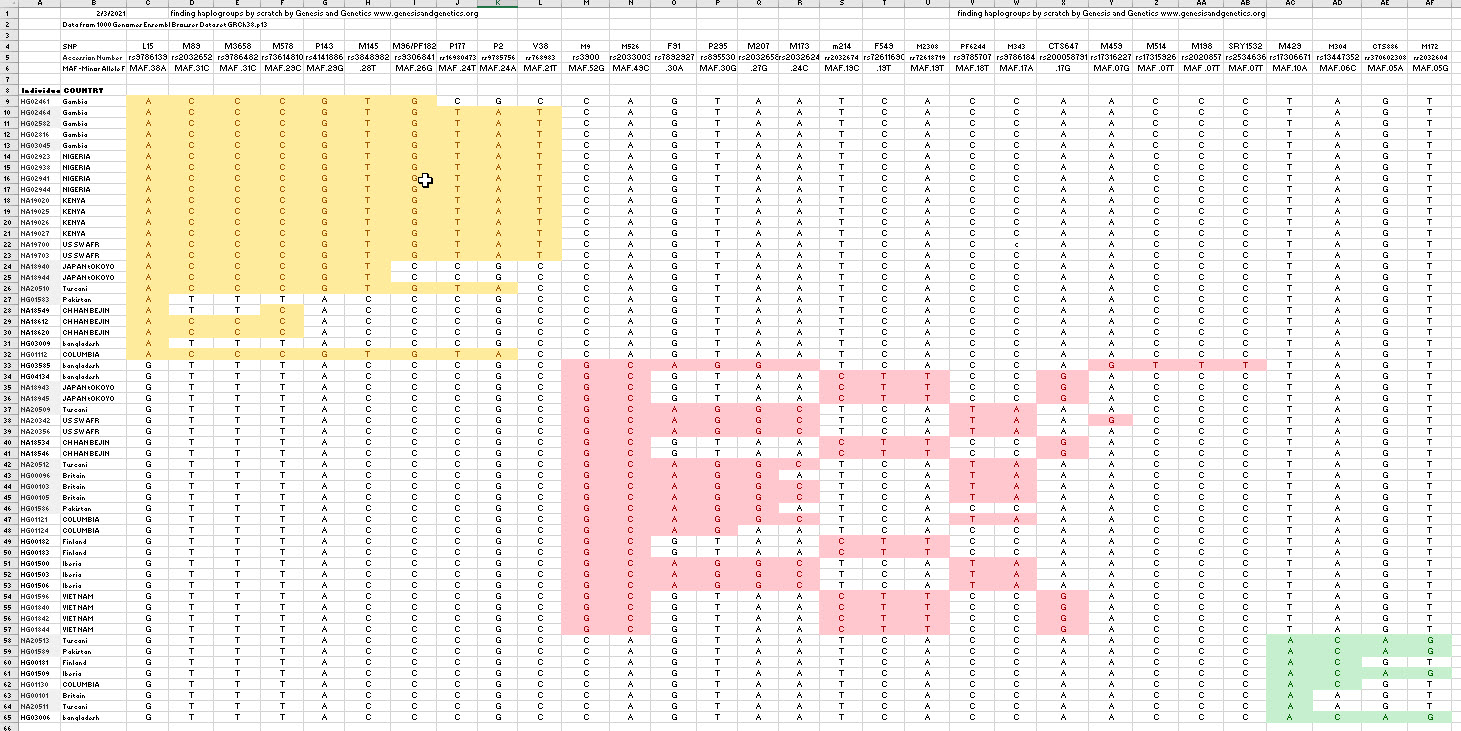
Next, we switched columns and rows to form groups. The groups were obvious. We also took each group and moved the columns so that the columns for each group with the largest MAF were on the left. Now we can see a clear pattern in the figure below. We see that each individual fits into just one group and had no mutations in any other group. Also, we see that every individual in the group has the mutation with the greatest MAF; this is the mutation of the most ancient ancestor of that group. Here is what the spreadsheet looks like after grouping: Note: you can CLICK HERE to see the matrix in EXCEL.
Looking at the matrix above, we see that each group has one maximum MAF. The yellow is .38, the red is 52, and the green is .10. They add up to 100 percent.
Now that we have shown that our most ancient ancestor had three sons, we can state that this finding is consistent with the Bible. The three groups are consistent with the three sons of Noah: Ham, Shem, and Japheth. The Bible states that, generally, those from Ham went south to Africa and southern Asia, those from Japheth went north to northern Europe and northern Asia, and those from Shem stayed in the Caucuses, the Near East, and North Africa. Therefore, one could easily conclude that Ham is the yellow group, Shem is the green group, and Japheth is the red group. Note: Noah’s Y-chromosome is also known since it would be that of the three sons with all mutations removed.
Bible in Genesis 10, the Table of Nations, states that all humanity came from these three sons:
Genesis 10:1 Now these are the generations of the sons of Noah, Shem, Ham, and Japheth: and unto them were sons born after the flood.
Genesis 10:32 These are the families of the sons of Noah, after their generations, in their nations: and by these were the nations divided in the earth after the flood.
This means that roughly 1/3 of the earth’s male population came from each son, but, since Japheth had 7 sons, Ham had 4 sons, and Shem had 5 sons; therefore, the worldwide distribution should be closer to the following:
Patriarch Number of sons % Projected World Population Observation
Japheth 7 sons 43.75 52
Ham 4 sons 25 38
Shem 5 sons 31.25 10
The chart above shows that both the Ham and Japheth results were higher than expected, and Shem was lower than expected. However, this can be explained in at least two ways:
(1) The Ham and Japheth offspring are in the world’s heavily populated areas, namely South and East Asia.
(2) The Thousand Genomes Project did not provide representatives of the Near East, the Mid-East, or Northern Africa, all of which would increase the Shem percentage, thereby decreasing Ham and Japheth’s percentages.
The above two issues are not meant as criticism but offered as an explanation.
Our findings are consistent with the Bible, secular ancient history, and genetic diversity:
- The Bible documents Noah and his family to be the only ones spared from the deluge which flooded the entire earth, and the earth was repopulated through Noah’s three sons.
- The historical period began when the Sumerians began writing cuneiform tablets. These early writings documented the kings before and after the flood. Also, the Sumerian legends are consistent with the world being populated by the three sons on the Ark.
- According to the Bible and Sumerian history, all humanity came from one family. As they migrated throughout the world, the genetic diversity would be lost from those who separated from the core population; therefore, the most genetic diversity should be where Noah’s family settled, the Near East. Those who migrated to the Americas, Australia, and southern Africa lost some genetic diversity. We can consider two levels of observing genetic diversity, first, what we see and, second, what DNA tells us. We can easily see eye color, skin color, and hair color. Looking at a globe, it is apparent that those with the most visual diversity meet in the Near East, the place of disembarkation from Noah’s Ark. From a DNA standpoint, one can look at the diversity of haplogroups, both Y-chromosomal and mitochondrial. The extremities are South Africa, Australia, and the Americas, which have only a few haplogroups. The location with the most haplogroups is, again, in the Near East.
To summarize our findings:
(1) From a Y-chromosome perspective, it appears that all humanity came from three male humans.
(2) Item (1) is not proof of the Biblical narrative concerning Noah’s three sons, but it is consistent with it. To prove it, one would have to sequence every human male ever born and analyze his genome. If anyone can trace an individual human Y Chromosome back to some basal mutation other than the three given, please let us know and comment below.
(3) We can now project Noah’s DNA; it is that of his three sons with no mutations.
(4) Our findings are consistent with the Bible, Sumerian history, and our current state of human diversity.
Future analysis It is now possible to know the Y-chromosome DNA of each of Noah’s grandchildren. This is in our job jar.
References:
2 https://www.internationalgenome.org/
3 https://useast.ensembl.org/index.html
Appendix A
If you would like to repeat this analysis or add to it, the following is the step-by-step procedure we used.
Let’s start by taking someone from, perhaps, Peru and find out to which group he belongs. Did he come from Shem, Ham, or Japheth?
(1) We open Ensembl (https://useast.ensembl.org/index.html ). Leave everything default and enter rs9786139 in the upper right-hand box—hit enter.
(2) You should get a new window with the following “rs9786139 (Human Variant).” Click on it. Notice that the MAF is .38.
(3) Click on the ICON that says, “population genetics.” Scroll down to find “AMR” and the subgroup “PEL.” Click on the “PEL.” You can see that 95 percent of this group has a “G,” and 5 percent of the population have an “A.” Next, go to the far right of the “PEL” row and click on “show.” Each of the rows of this screen is an actual human being. You will notice that most of this group has a “G,” but a few have an “A.”
(4) Looking at the population genetics of those from PEL, Peru, you can see that a few Peruvians are from Ham, but most are from someone else. You can find out who by entering the defining mutations for Shem and Japheth. If you enter all the mutations and analyze the same Peruvian, you will see all his mutations are in one group or another. If you look at rs3900, you will see that most Peruvians are from Japheth.
(5) continue entering the mutations one by one, and you will see that our Peruvian is only from one group and has no mutations from any other group. If you do a significant number of individuals, please sent them to us.

Jepheth’s sons went in 3 directions
(east, west & north) right? Then grandsons, great grandsons.. for generations spread.
Can’t geneticists follow Y haplogroups to specific areas/countries?
If rule is that haplogroup is passed on, then
WHY are there so many different haplogroups?
Ann,
Every time there is a new mutation, a new haplogroup is defined. If your father had a new mutation, he would define a new haplogroup, it would be a very small haplogroup. But, in 500 years, it would be a bigger haplogroup. Now the world has approximately 4 billion men. So, each new haplogroup is one-four-billionth of the world population. When the world’s male population reaches higher populations, your father’s haplogroup will continue to be one-four-billionth of the world’s male population, on average.
The mutations in 1 out of a thousand males were formed when the population was approximately 1000 males. And that generation of males formed probably more than 1000 haplogroups. In our analysis, we considered only haplogroups in at least 5 percent of the population. These are mainly the grandsons of Noah.
In summary, there are many new haplogroups, but they are subgroups of our ancient ancestor’s haplogroups.